One of the earliest applications of predictive analytics methods applied so far was to automatically recognize letters, which post office machines use to sort mail. In this analysis, I’ll build a model that uses statistics of images of four letters in the Roman alphabet – A, B, P, and R – to predict which letter a particular image corresponds to.
Note, that this is a multiclass classification problem. We have mostly focused on binary classification problems (e.g., predicting whether an individual voted or not, whether the Supreme Court will affirm or reverse a case, whether or not a person is at risk for a certain disease, etc.). In this problem, we have more than two classifications that are possible for each observation.
The file letters_ABPR.csv contains 3116 observations, each of which corresponds to a certain image of one of the four letters A, B, P and R. The images came from 20 different fonts, which were then randomly distorted to produce the final images; each such distorted image is represented as a collection of pixels, each of which is “on” or “off”.
For each such distorted image, we have available certain statistics of the image in terms of these pixels, as well as which of the four letters the image is. This data comes from the UCI ML Repository.
This dataset contains the following 17 variables:
- letter = the letter that the image corresponds to (A, B, P or R)
- xbox = the horizontal position of where the smallest box covering the letter shape begins.
- ybox = the vertical position of where the smallest box covering the letter shape begins.
- width = the width of this smallest box.
- height = the height of this smallest box.
- onpix = the total number of “on” pixels in the character image
- xbar = the mean horizontal position of all of the “on” pixels
- ybar = the mean vertical position of all of the “on” pixels
- x2bar = the mean squared horizontal position of all of the “on” pixels in the image
- y2bar = the mean squared vertical position of all of the “on” pixels in the image
- xybar = the mean of the product of the horizontal and vertical position of all of the “on” pixels in the image
- x2ybar = the mean of the product of the squared horizontal position and the vertical position of all of the “on” pixels
- xy2bar = the mean of the product of the horizontal position and the squared vertical position of all of the “on” pixels
- xedge = the mean number of edges (the number of times an “off” pixel is followed by an “on” pixel, or the image boundary is hit) as the image is scanned from left to right, along the whole vertical length of the image
- xedgeycor = the mean of the product of the number of horizontal edges at each vertical position and the vertical position
- yedge = the mean number of edges as the images is scanned from top to bottom, along the whole horizontal length of the image
- yedgexcor = the mean of the product of the number of vertical edges at each horizontal position and the horizontal position
Load the dataset
letters <- read.csv("letters_ABPR.csv")
summary(letters) letter xbox ybox width
A:789 Min. : 0.000 Min. : 0.000 Min. : 1.000
B:766 1st Qu.: 3.000 1st Qu.: 5.000 1st Qu.: 4.000
P:803 Median : 4.000 Median : 7.000 Median : 5.000
R:758 Mean : 3.915 Mean : 7.051 Mean : 5.186
3rd Qu.: 5.000 3rd Qu.: 9.000 3rd Qu.: 6.000
Max. :13.000 Max. :15.000 Max. :11.000
height onpix xbar ybar
Min. : 0.000 Min. : 0.000 Min. : 3.000 Min. : 0.000
1st Qu.: 4.000 1st Qu.: 2.000 1st Qu.: 6.000 1st Qu.: 6.000
Median : 6.000 Median : 4.000 Median : 7.000 Median : 7.000
Mean : 5.276 Mean : 3.869 Mean : 7.469 Mean : 7.197
3rd Qu.: 7.000 3rd Qu.: 5.000 3rd Qu.: 8.000 3rd Qu.: 9.000
Max. :12.000 Max. :12.000 Max. :14.000 Max. :15.000
x2bar y2bar xybar x2ybar
Min. : 0.000 Min. :0.000 Min. : 3.000 Min. : 0.00
1st Qu.: 3.000 1st Qu.:2.000 1st Qu.: 7.000 1st Qu.: 3.00
Median : 4.000 Median :4.000 Median : 8.000 Median : 5.00
Mean : 4.706 Mean :3.903 Mean : 8.491 Mean : 4.52
3rd Qu.: 6.000 3rd Qu.:5.000 3rd Qu.:10.000 3rd Qu.: 6.00
Max. :11.000 Max. :8.000 Max. :14.000 Max. :10.00
xy2bar xedge xedgeycor yedge
Min. : 0.000 Min. : 0.000 Min. : 1.000 Min. : 0.0
1st Qu.: 6.000 1st Qu.: 2.000 1st Qu.: 7.000 1st Qu.: 3.0
Median : 7.000 Median : 2.000 Median : 8.000 Median : 4.0
Mean : 6.711 Mean : 2.913 Mean : 7.763 Mean : 4.6
3rd Qu.: 8.000 3rd Qu.: 4.000 3rd Qu.: 9.000 3rd Qu.: 6.0
Max. :14.000 Max. :10.000 Max. :13.000 Max. :12.0
yedgexcor
Min. : 1.000
1st Qu.: 7.000
Median : 8.000
Mean : 8.418
3rd Qu.:10.000
Max. :13.000 str(letters)'data.frame': 3116 obs. of 17 variables:
$ letter : Factor w/ 4 levels "A","B","P","R": 2 1 4 2 3 4 4 1 3 3 ...
$ xbox : int 4 1 5 5 3 8 2 3 8 6 ...
$ ybox : int 2 1 9 9 6 10 6 7 14 10 ...
$ width : int 5 3 5 7 4 8 4 5 7 8 ...
$ height : int 4 2 7 7 4 6 4 5 8 8 ...
$ onpix : int 4 1 6 10 2 6 3 3 4 7 ...
$ xbar : int 8 8 6 9 4 7 6 12 5 8 ...
$ ybar : int 7 2 11 8 14 7 7 2 10 5 ...
$ x2bar : int 6 2 7 4 8 3 5 3 6 7 ...
$ y2bar : int 6 2 3 4 1 5 5 2 3 5 ...
$ xybar : int 7 8 7 6 11 8 6 10 12 7 ...
$ x2ybar : int 6 2 3 8 6 4 5 2 5 6 ...
$ xy2bar : int 6 8 9 6 3 8 7 9 4 6 ...
$ xedge : int 2 1 2 6 0 6 3 2 4 3 ...
$ xedgeycor: int 8 6 7 11 10 6 7 6 10 9 ...
$ yedge : int 7 2 5 8 4 7 5 3 4 8 ...
$ yedgexcor: int 10 7 11 7 8 7 8 8 8 9 ...Problem 1.1 - Predicting B or not B
Let’s warm up by attempting to predict just whether a letter is B or not. To begin, load the file letters_ABPR.csv into R, and call it letters. Then, create a new variable isB in the dataframe, which takes the value “TRUE” if the observation corresponds to the letter B, and “FALSE” if it does not.
letters$isB <- as.factor(letters$letter == "B")Now, split the dataset into a training and testing set, putting 50% of the data in the training set. Set the seed to 1000 before making the split. The first argument to sample.split should be the dependent variable “letters$isB”. Remember that TRUE values from sample.split should go in the training set.
library(caTools)
set.seed(1000)
lettersSplit = sample.split(letters$isB, SplitRatio = 0.5)
lettersTrain = subset(letters, lettersSplit == TRUE)
lettersTest = subset(letters, lettersSplit == FALSE)Before building models, let’s consider a baseline method that always predicts the most frequent outcome, which is “not B”.
What is the accuracy of this baseline method on the test set?
table(letters$isB)
FALSE TRUE
2350 766 table(lettersTest$isB)
FALSE TRUE
1175 383 1175 / (1175 + 383)[1] 0.754172Problem 1.2 - Predicting B or not B
Now, build a classification tree to predict whether a letter is a B or not, using the training set to build the model. Remember to remove the variable “letter” out of the model, as this is related to what we are trying to predict!
library(rpart)
library(rpart.plot)
CARTb <- rpart(isB ~ . - letter, data = lettersTrain, method="class")We are just using the default parameters in our CART model, so we don’t need to add the minbucket or cp arguments at all. We also added the argument method=“class” since this is a classification problem.
What is the accuracy of the CART model on the test-set? (Use type=“class” when making predictions on the test set.)
bPredict <- predict(CARTb, newdata = lettersTest, type = "class")
table(lettersTest$isB, bPredict) bPredict
FALSE TRUE
FALSE 1118 57
TRUE 43 340(1126 + 342) / nrow(lettersTest)[1] 0.9422336Problem 1.3 - Predicting B or Not B
Now, build a random forest model to predict whether the letter is a B or not (the isB variable) using the training set. Using all of the other variables as independent variables, except letter (since it helped us define what we are trying to predict!). Using the default settings for ntree and nodesize (don’t include these arguments at all). Right before building the model, set the seed to 1000. (NOTE: You might get a slightly different answer on this problem, even if you set the random seed. This has to do with your operating system and the implementation of the random forest algorithm.)
What is the accuracy of the model on the test set?
bForestPredict <- predict(bForest, newdata = lettersTest, type = "class")
table(lettersTest$isB, bForestPredict) bForestPredict
FALSE TRUE
FALSE 1163 12
TRUE 9 374(1164 + 372) / nrow(lettersTest)[1] 0.9858793Random forests tends to improve on CART in terms of predictive accuracy. Sometimes, this improvement can be quite significant, as it is here.
Problem 2.1 - Predicting the letters A, B, P, R
Let us now move on to the problem that we were originally interested in, which is to predict whether or not a letter is one of the four letters A, B, P or R.
As we saw earlier, building a multiclass classification CART model is no harder than building the models for binary classification problems. Fortunately, building a random forest model is just as easy. The variable in our dataframe which we will be trying to predict is “letter”.
Start by converting letter in the original dataset (letters) to a factor by running the following code:
letters$letter <- as.factor(letters$letter)Now, generate new training and testing sets of the letters dataframe using letters$letter as the first input to the sample.split function. Before splitting, set your seed to 2000. Again put 50% of the data in the training set. (Why do we need to split the data again? Remember that sample.split balances the outcome variable in the training and testing sets. With a new outcome variable, we want to re-generate our split.)
set.seed(2000)
lettersAllSplit <- sample.split(letters$letter, SplitRatio = 0.5)
lettersAllTrain <- subset(letters, lettersAllSplit == TRUE)
lettersAllTest <- subset(letters, lettersAllSplit == FALSE)In a multiclass classification problem, a simple baseline model is to predict the most frequent class of all of the options.
What is the baseline accuracy on the testing set?
table(letters$letter)
A B P R
789 766 803 758 table(lettersAllTest$letter)
A B P R
395 383 401 379 401 / nrow(lettersAllTest)[1] 0.2573813P is the most frequent class in the test set
Problem 2.2 - Predicting the letters A, B, P, R
Now build a classification tree to predict “letter”, using the training set to build our model. You should use all of the other variables as independent variables, except “isB”, since it is related to what we are trying to predict!
Just use the default parameters in your CART model. Add the argument method=“class” since this is a classification problem. Even though we have multiple classes here, nothing changes in how we build the model from the binary case.
CARTletters <- rpart(letter ~ . - isB, data = lettersAllTrain, method="class")
summary(CARTletters)Call:
rpart(formula = letter ~ . - isB, data = lettersAllTrain, method = "class")
n= 1558
CP nsplit rel error xerror xstd
1 0.31920415 0 1.0000000 1.0346021 0.01442043
2 0.25865052 1 0.6807958 0.6323529 0.01704001
3 0.18685121 2 0.4221453 0.4238754 0.01585412
4 0.02595156 3 0.2352941 0.2370242 0.01299919
5 0.02076125 4 0.2093426 0.2162630 0.01253234
6 0.01730104 5 0.1885813 0.1980969 0.01209034
7 0.01384083 6 0.1712803 0.1894464 0.01186782
8 0.01211073 7 0.1574394 0.1678201 0.01127370
9 0.01000000 8 0.1453287 0.1608997 0.01107113
Variable importance
ybar xedgeycor x2ybar xy2bar yedge y2bar xedge
17 16 14 12 11 8 7
xybar x2bar xbar
5 5 3
Node number 1: 1558 observations, complexity param=0.3192042
predicted class=P expected loss=0.7419769 P(node) =1
class counts: 394 383 402 379
probabilities: 0.253 0.246 0.258 0.243
left son=2 (1088 obs) right son=3 (470 obs)
Primary splits:
xedgeycor < 8.5 to the left, improve=293.2010, (0 missing)
ybar < 5.5 to the left, improve=287.8322, (0 missing)
xy2bar < 5.5 to the right, improve=278.1742, (0 missing)
x2ybar < 2.5 to the left, improve=262.6356, (0 missing)
yedge < 4.5 to the left, improve=177.0582, (0 missing)
Surrogate splits:
xy2bar < 5.5 to the right, agree=0.892, adj=0.643, (0 split)
ybar < 8.5 to the left, agree=0.821, adj=0.406, (0 split)
xedge < 1.5 to the right, agree=0.816, adj=0.391, (0 split)
xybar < 10.5 to the left, agree=0.785, adj=0.287, (0 split)
x2ybar < 6.5 to the left, agree=0.777, adj=0.262, (0 split)
Node number 2: 1088 observations, complexity param=0.2586505
predicted class=A expected loss=0.6488971 P(node) =0.6983312
class counts: 382 338 13 355
probabilities: 0.351 0.311 0.012 0.326
left son=4 (344 obs) right son=5 (744 obs)
Primary splits:
ybar < 5.5 to the left, improve=275.7625, (0 missing)
x2ybar < 2.5 to the left, improve=240.6702, (0 missing)
y2bar < 2.5 to the left, improve=226.4519, (0 missing)
yedge < 3.5 to the left, improve=215.2610, (0 missing)
xedgeycor < 7.5 to the right, improve=171.4917, (0 missing)
Surrogate splits:
x2ybar < 2.5 to the left, agree=0.904, adj=0.698, (0 split)
y2bar < 2.5 to the left, agree=0.892, adj=0.657, (0 split)
yedge < 3.5 to the left, agree=0.881, adj=0.625, (0 split)
x2bar < 2.5 to the left, agree=0.820, adj=0.430, (0 split)
xbar < 9.5 to the right, agree=0.779, adj=0.302, (0 split)
Node number 3: 470 observations, complexity param=0.01730104
predicted class=P expected loss=0.1723404 P(node) =0.3016688
class counts: 12 45 389 24
probabilities: 0.026 0.096 0.828 0.051
left son=6 (91 obs) right son=7 (379 obs)
Primary splits:
xybar < 7.5 to the left, improve=59.48719, (0 missing)
xy2bar < 6.5 to the right, improve=54.86112, (0 missing)
ybar < 7.5 to the left, improve=49.49367, (0 missing)
yedge < 6.5 to the right, improve=48.42295, (0 missing)
xedge < 5.5 to the left, improve=30.83057, (0 missing)
Surrogate splits:
xy2bar < 6.5 to the right, agree=0.936, adj=0.670, (0 split)
ybar < 7.5 to the left, agree=0.902, adj=0.495, (0 split)
xedge < 5.5 to the right, agree=0.889, adj=0.429, (0 split)
yedge < 6.5 to the right, agree=0.885, adj=0.407, (0 split)
onpix < 6.5 to the right, agree=0.838, adj=0.165, (0 split)
Node number 4: 344 observations
predicted class=A expected loss=0.04360465 P(node) =0.2207959
class counts: 329 9 3 3
probabilities: 0.956 0.026 0.009 0.009
Node number 5: 744 observations, complexity param=0.1868512
predicted class=R expected loss=0.5268817 P(node) =0.4775353
class counts: 53 329 10 352
probabilities: 0.071 0.442 0.013 0.473
left son=10 (342 obs) right son=11 (402 obs)
Primary splits:
xedgeycor < 7.5 to the right, improve=139.70670, (0 missing)
xy2bar < 7.5 to the left, improve= 92.43059, (0 missing)
x2ybar < 5.5 to the right, improve= 81.07422, (0 missing)
y2bar < 4.5 to the right, improve= 56.45671, (0 missing)
yedgexcor < 10.5 to the left, improve= 52.58754, (0 missing)
Surrogate splits:
x2ybar < 5.5 to the right, agree=0.738, adj=0.430, (0 split)
xy2bar < 6.5 to the left, agree=0.675, adj=0.292, (0 split)
xedge < 2.5 to the left, agree=0.675, adj=0.292, (0 split)
yedge < 5.5 to the right, agree=0.644, adj=0.225, (0 split)
ybar < 7.5 to the left, agree=0.625, adj=0.184, (0 split)
Node number 6: 91 observations, complexity param=0.01384083
predicted class=B expected loss=0.5604396 P(node) =0.05840822
class counts: 10 40 20 21
probabilities: 0.110 0.440 0.220 0.231
left son=12 (55 obs) right son=13 (36 obs)
Primary splits:
x2bar < 3.5 to the right, improve=14.308240, (0 missing)
xy2bar < 7.5 to the left, improve= 9.472092, (0 missing)
yedge < 4.5 to the left, improve= 9.449763, (0 missing)
x2ybar < 7.5 to the right, improve= 8.053076, (0 missing)
yedgexcor < 6.5 to the right, improve= 7.478284, (0 missing)
Surrogate splits:
yedgexcor < 5.5 to the right, agree=0.736, adj=0.333, (0 split)
x2ybar < 7.5 to the left, agree=0.725, adj=0.306, (0 split)
yedge < 5.5 to the right, agree=0.725, adj=0.306, (0 split)
xy2bar < 8.5 to the left, agree=0.714, adj=0.278, (0 split)
ybar < 7.5 to the left, agree=0.681, adj=0.194, (0 split)
Node number 7: 379 observations
predicted class=P expected loss=0.02638522 P(node) =0.2432606
class counts: 2 5 369 3
probabilities: 0.005 0.013 0.974 0.008
Node number 10: 342 observations, complexity param=0.02595156
predicted class=B expected loss=0.2192982 P(node) =0.2195122
class counts: 14 267 10 51
probabilities: 0.041 0.781 0.029 0.149
left son=20 (283 obs) right son=21 (59 obs)
Primary splits:
xy2bar < 7.5 to the left, improve=48.65030, (0 missing)
xedge < 2.5 to the left, improve=33.98799, (0 missing)
y2bar < 4.5 to the right, improve=27.13499, (0 missing)
yedgexcor < 6.5 to the left, improve=15.49245, (0 missing)
ybar < 8.5 to the left, improve=15.03303, (0 missing)
Surrogate splits:
xedge < 5.5 to the left, agree=0.871, adj=0.254, (0 split)
yedgexcor < 4.5 to the right, agree=0.854, adj=0.153, (0 split)
ybar < 9.5 to the left, agree=0.848, adj=0.119, (0 split)
xbox < 6.5 to the left, agree=0.842, adj=0.085, (0 split)
ybox < 11.5 to the left, agree=0.842, adj=0.085, (0 split)
Node number 11: 402 observations, complexity param=0.02076125
predicted class=R expected loss=0.2512438 P(node) =0.2580231
class counts: 39 62 0 301
probabilities: 0.097 0.154 0.000 0.749
left son=22 (26 obs) right son=23 (376 obs)
Primary splits:
yedge < 2.5 to the left, improve=35.46191, (0 missing)
x2ybar < 0.5 to the left, improve=34.14932, (0 missing)
y2bar < 1.5 to the left, improve=33.87850, (0 missing)
x2bar < 3.5 to the left, improve=19.57685, (0 missing)
yedgexcor < 8.5 to the left, improve=19.07812, (0 missing)
Surrogate splits:
y2bar < 1.5 to the left, agree=0.993, adj=0.885, (0 split)
x2ybar < 0.5 to the left, agree=0.993, adj=0.885, (0 split)
Node number 12: 55 observations
predicted class=B expected loss=0.3090909 P(node) =0.03530167
class counts: 1 38 13 3
probabilities: 0.018 0.691 0.236 0.055
Node number 13: 36 observations
predicted class=R expected loss=0.5 P(node) =0.02310655
class counts: 9 2 7 18
probabilities: 0.250 0.056 0.194 0.500
Node number 20: 283 observations
predicted class=B expected loss=0.08480565 P(node) =0.1816431
class counts: 3 259 8 13
probabilities: 0.011 0.915 0.028 0.046
Node number 21: 59 observations
predicted class=R expected loss=0.3559322 P(node) =0.03786906
class counts: 11 8 2 38
probabilities: 0.186 0.136 0.034 0.644
Node number 22: 26 observations
predicted class=A expected loss=0.03846154 P(node) =0.01668806
class counts: 25 0 0 1
probabilities: 0.962 0.000 0.000 0.038
Node number 23: 376 observations, complexity param=0.01211073
predicted class=R expected loss=0.2021277 P(node) =0.241335
class counts: 14 62 0 300
probabilities: 0.037 0.165 0.000 0.798
left son=46 (26 obs) right son=47 (350 obs)
Primary splits:
yedge < 7.5 to the right, improve=19.73450, (0 missing)
x2ybar < 5.5 to the right, improve=16.32647, (0 missing)
xybar < 8.5 to the right, improve=15.20779, (0 missing)
xedge < 3.5 to the right, improve=14.35240, (0 missing)
onpix < 4.5 to the right, improve=12.94437, (0 missing)
Surrogate splits:
xedgeycor < 4.5 to the left, agree=0.939, adj=0.115, (0 split)
Node number 46: 26 observations
predicted class=B expected loss=0.3076923 P(node) =0.01668806
class counts: 4 18 0 4
probabilities: 0.154 0.692 0.000 0.154
Node number 47: 350 observations
predicted class=R expected loss=0.1542857 P(node) =0.224647
class counts: 10 44 0 296
probabilities: 0.029 0.126 0.000 0.846 prp(CARTletters)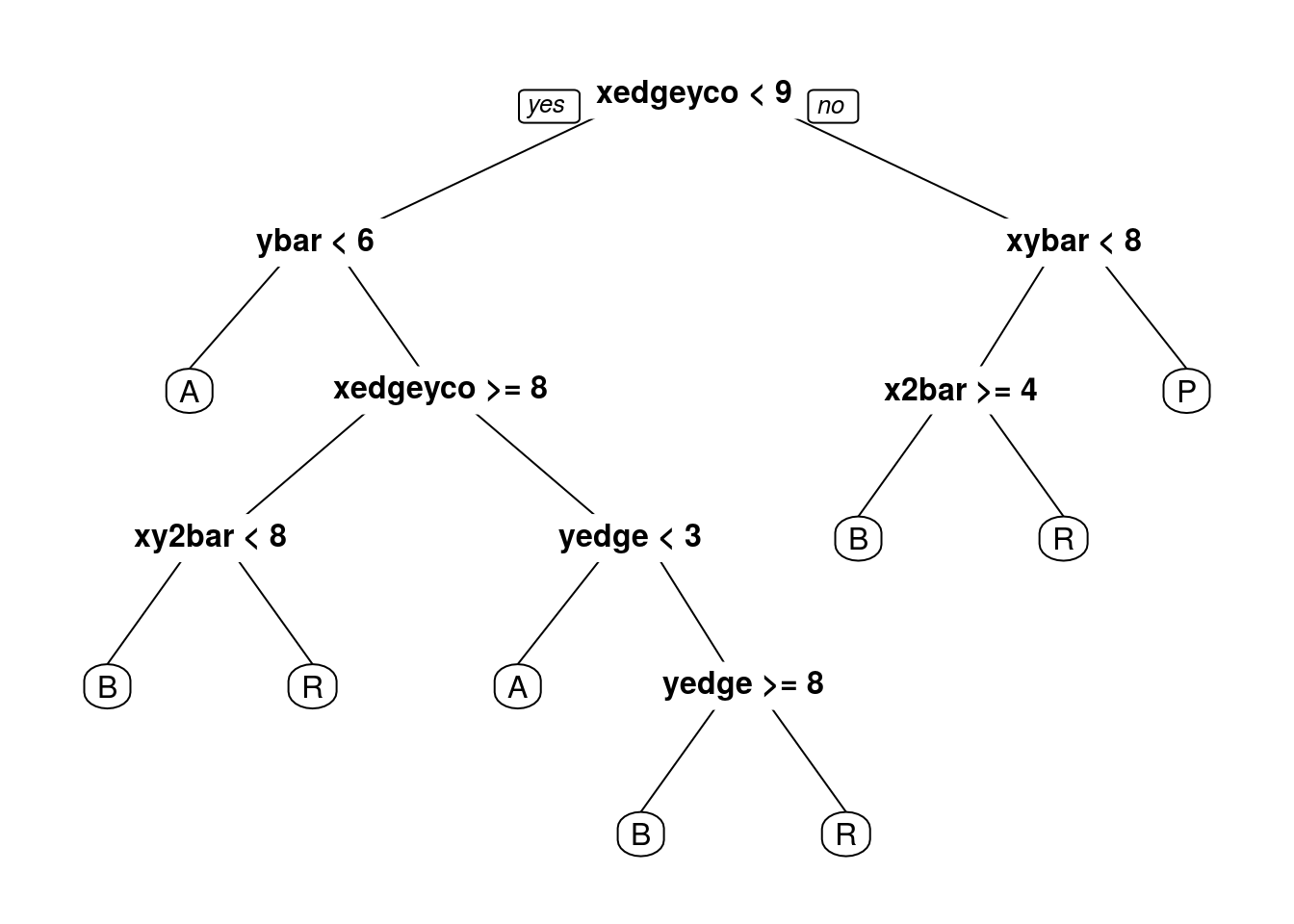
What is the test-set accuracy of our CART model? Use the argument type=“class” when making predictions. (HINT: When you are computing the test-set accuracy using the confusion matrix, you want to add everything on the main diagonal and divide by the total number of observations in the test-set, which can be computed with nrow(test), where test is the name of our test-set).
lettersPredict <-
as.vector(predict(CARTletters, newdata = lettersAllTest, type = "class"))
length(lettersPredict)[1] 1558lettersPredict [1] "B" "R" "A" "P" "A" "R" "A" "A" "A" "B" "A" "A" "P" "P" "B" "R" "B"
[18] "P" "P" "B" "A" "A" "B" "P" "R" "R" "A" "A" "B" "P" "A" "B" "P" "B"
[35] "B" "A" "P" "B" "B" "R" "A" "R" "R" "R" "R" "B" "B" "B" "P" "R" "P"
[52] "B" "R" "R" "B" "B" "R" "A" "R" "P" "R" "P" "B" "R" "B" "A" "P" "P"
[69] "R" "R" "A" "B" "P" "R" "R" "R" "A" "P" "R" "R" "P" "B" "B" "R" "A"
[86] "P" "A" "A" "A" "P" "A" "A" "A" "B" "B" "B" "R" "R" "P" "P" "B" "A"
[103] "R" "P" "R" "R" "B" "A" "P" "P" "R" "P" "A" "A" "P" "R" "P" "R" "P"
[120] "B" "R" "R" "R" "B" "R" "A" "P" "A" "B" "P" "P" "P" "B" "R" "A" "P"
[137] "P" "R" "B" "R" "R" "A" "B" "A" "P" "R" "R" "A" "R" "A" "B" "B" "B"
[154] "B" "B" "P" "A" "P" "R" "R" "A" "A" "P" "A" "A" "P" "B" "B" "B" "P"
[171] "P" "B" "R" "B" "R" "P" "B" "P" "B" "A" "P" "A" "A" "B" "R" "P" "P"
[188] "P" "R" "A" "B" "B" "A" "R" "B" "R" "P" "R" "A" "R" "A" "A" "R" "R"
[205] "B" "R" "B" "A" "B" "P" "R" "A" "A" "R" "B" "R" "A" "R" "P" "R" "R"
[222] "B" "P" "P" "A" "A" "B" "P" "B" "P" "R" "P" "A" "R" "R" "R" "A" "A"
[239] "B" "P" "R" "R" "A" "R" "A" "P" "B" "R" "P" "B" "A" "R" "B" "A" "A"
[256] "A" "B" "B" "A" "P" "A" "B" "P" "R" "A" "R" "P" "B" "P" "R" "P" "A"
[273] "R" "R" "R" "B" "R" "R" "P" "R" "P" "P" "A" "R" "P" "R" "R" "A" "P"
[290] "B" "A" "A" "A" "A" "B" "R" "R" "A" "P" "R" "B" "P" "A" "R" "R" "R"
[307] "R" "A" "B" "B" "R" "P" "R" "B" "A" "A" "P" "R" "A" "B" "P" "A" "R"
[324] "B" "R" "P" "P" "A" "P" "R" "A" "R" "A" "B" "P" "R" "P" "R" "P" "B"
[341] "B" "B" "A" "A" "B" "B" "B" "P" "P" "P" "B" "B" "A" "P" "R" "R" "R"
[358] "R" "P" "B" "P" "B" "P" "R" "B" "P" "B" "R" "B" "A" "P" "R" "R" "P"
[375] "B" "B" "R" "A" "R" "A" "R" "R" "A" "B" "P" "R" "A" "P" "P" "A" "A"
[392] "A" "P" "R" "R" "P" "B" "A" "A" "R" "P" "A" "R" "B" "A" "P" "P" "P"
[409] "B" "A" "R" "P" "R" "P" "A" "A" "R" "P" "P" "R" "R" "R" "B" "R" "P"
[426] "B" "A" "R" "R" "P" "P" "A" "A" "B" "A" "P" "P" "B" "P" "A" "R" "B"
[443] "P" "B" "R" "P" "A" "A" "B" "P" "B" "R" "A" "P" "A" "R" "P" "P" "P"
[460] "R" "B" "B" "P" "P" "R" "B" "R" "B" "P" "R" "B" "A" "R" "B" "P" "B"
[477] "R" "R" "P" "R" "B" "B" "A" "P" "B" "P" "P" "B" "A" "A" "A" "B" "P"
[494] "P" "R" "B" "A" "A" "B" "A" "R" "R" "R" "B" "R" "B" "B" "R" "R" "A"
[511] "A" "P" "R" "P" "A" "R" "B" "A" "P" "P" "R" "P" "B" "B" "A" "A" "P"
[528] "A" "B" "P" "A" "P" "A" "R" "A" "A" "R" "R" "P" "R" "A" "R" "R" "R"
[545] "B" "B" "R" "R" "R" "R" "A" "B" "R" "P" "B" "A" "A" "R" "R" "R" "R"
[562] "R" "R" "P" "A" "P" "R" "A" "R" "R" "R" "B" "R" "B" "B" "A" "R" "P"
[579] "B" "R" "B" "R" "P" "B" "A" "A" "B" "R" "A" "A" "B" "P" "P" "B" "B"
[596] "B" "P" "R" "A" "P" "P" "B" "B" "P" "B" "R" "P" "P" "P" "R" "A" "A"
[613] "B" "P" "B" "B" "R" "B" "A" "B" "A" "P" "R" "R" "A" "P" "B" "A" "P"
[630] "A" "P" "R" "R" "P" "B" "R" "A" "A" "A" "B" "B" "P" "B" "A" "P" "P"
[647] "P" "B" "A" "P" "P" "R" "P" "P" "R" "R" "B" "R" "A" "P" "B" "P" "P"
[664] "A" "P" "A" "R" "P" "P" "R" "A" "R" "P" "A" "B" "B" "R" "P" "P" "R"
[681] "A" "P" "B" "P" "R" "P" "A" "A" "A" "R" "A" "P" "R" "A" "R" "P" "P"
[698] "A" "B" "R" "R" "R" "R" "P" "R" "B" "R" "P" "B" "R" "B" "B" "B" "P"
[715] "A" "B" "P" "R" "B" "R" "B" "R" "B" "R" "B" "P" "A" "A" "R" "A" "A"
[732] "P" "R" "P" "P" "P" "P" "P" "R" "R" "R" "A" "P" "P" "B" "B" "A" "A"
[749] "B" "B" "R" "R" "B" "B" "R" "A" "B" "P" "P" "R" "A" "R" "P" "B" "B"
[766] "P" "P" "R" "B" "R" "B" "P" "B" "A" "A" "B" "A" "P" "A" "R" "R" "B"
[783] "R" "R" "R" "R" "P" "P" "P" "R" "R" "P" "A" "B" "B" "B" "A" "A" "P"
[800] "A" "R" "P" "R" "B" "B" "R" "P" "B" "R" "R" "R" "A" "B" "R" "R" "A"
[817] "B" "P" "P" "P" "B" "R" "P" "A" "R" "A" "A" "R" "R" "P" "P" "R" "R"
[834] "A" "P" "P" "B" "R" "P" "B" "A" "A" "R" "A" "R" "B" "R" "R" "P" "B"
[851] "R" "P" "A" "P" "A" "A" "R" "A" "P" "R" "B" "B" "B" "R" "R" "P" "A"
[868] "P" "R" "R" "P" "R" "R" "B" "P" "R" "B" "B" "P" "P" "R" "B" "B" "P"
[885] "A" "R" "A" "A" "A" "R" "A" "B" "R" "B" "P" "B" "R" "B" "B" "B" "R"
[902] "B" "P" "P" "B" "B" "R" "R" "A" "B" "B" "B" "R" "P" "P" "R" "R" "A"
[919] "R" "P" "R" "P" "R" "B" "R" "B" "R" "P" "P" "R" "A" "R" "B" "A" "R"
[936] "B" "P" "A" "A" "B" "P" "A" "R" "B" "P" "R" "B" "R" "A" "P" "A" "R"
[953] "P" "R" "B" "B" "A" "A" "P" "B" "B" "B" "A" "P" "P" "B" "A" "R" "A"
[970] "P" "P" "R" "P" "R" "R" "A" "B" "R" "P" "P" "B" "A" "A" "P" "B" "B"
[987] "A" "A" "A" "A" "A" "R" "A" "P" "P" "A" "A" "A" "R" "B" "B" "P" "R"
[1004] "P" "A" "P" "B" "B" "B" "A" "B" "A" "P" "R" "A" "P" "R" "P" "A" "R"
[1021] "P" "A" "B" "R" "P" "R" "A" "P" "P" "P" "P" "B" "R" "A" "R" "P" "A"
[1038] "R" "A" "B" "A" "P" "R" "B" "A" "A" "R" "B" "P" "A" "B" "P" "B" "R"
[1055] "B" "A" "A" "B" "B" "R" "R" "R" "B" "P" "B" "P" "A" "P" "A" "A" "P"
[1072] "R" "B" "A" "P" "R" "R" "A" "A" "P" "R" "R" "R" "B" "R" "A" "R" "A"
[1089] "P" "B" "B" "B" "P" "B" "B" "R" "A" "B" "A" "P" "R" "A" "B" "B" "R"
[1106] "B" "B" "P" "B" "B" "R" "P" "P" "A" "P" "A" "B" "R" "A" "A" "R" "R"
[1123] "B" "A" "P" "B" "R" "A" "R" "B" "B" "A" "R" "P" "B" "P" "B" "B" "A"
[1140] "A" "P" "R" "R" "A" "R" "R" "P" "R" "A" "R" "R" "P" "B" "P" "R" "B"
[1157] "P" "A" "A" "P" "R" "P" "A" "R" "B" "R" "P" "A" "B" "P" "R" "B" "B"
[1174] "A" "R" "A" "B" "R" "R" "B" "A" "R" "A" "B" "R" "R" "R" "A" "A" "B"
[1191] "R" "B" "B" "P" "A" "R" "P" "A" "A" "R" "B" "P" "R" "P" "B" "A" "P"
[1208] "P" "P" "R" "R" "R" "P" "P" "R" "A" "P" "R" "B" "A" "B" "R" "P" "A"
[1225] "R" "P" "R" "R" "B" "A" "A" "P" "R" "P" "A" "A" "A" "B" "A" "P" "A"
[1242] "B" "B" "A" "B" "B" "R" "R" "P" "R" "B" "R" "P" "B" "A" "A" "R" "P"
[1259] "B" "A" "R" "R" "A" "R" "P" "P" "B" "A" "P" "R" "B" "A" "A" "P" "A"
[1276] "B" "R" "P" "P" "A" "P" "P" "B" "A" "A" "R" "P" "R" "A" "P" "R" "A"
[1293] "R" "R" "A" "P" "B" "A" "R" "B" "R" "R" "R" "B" "B" "R" "R" "A" "R"
[1310] "A" "R" "R" "P" "A" "R" "R" "A" "R" "B" "B" "B" "R" "R" "A" "A" "B"
[1327] "B" "B" "R" "P" "R" "R" "B" "R" "B" "A" "R" "A" "A" "A" "R" "B" "P"
[1344] "P" "P" "B" "B" "A" "P" "R" "A" "R" "P" "P" "P" "A" "R" "B" "A" "A"
[1361] "B" "B" "A" "P" "R" "B" "P" "P" "R" "R" "B" "R" "P" "B" "P" "A" "A"
[1378] "R" "B" "P" "P" "R" "P" "P" "P" "P" "A" "B" "R" "B" "A" "B" "R" "B"
[1395] "B" "P" "A" "A" "R" "R" "A" "B" "R" "P" "B" "R" "A" "P" "R" "B" "R"
[1412] "R" "B" "R" "R" "P" "P" "P" "R" "B" "A" "A" "A" "B" "P" "A" "B" "B"
[1429] "P" "B" "R" "R" "B" "A" "P" "B" "R" "R" "A" "P" "P" "P" "P" "P" "R"
[1446] "A" "B" "A" "B" "A" "B" "B" "P" "B" "A" "B" "P" "R" "B" "P" "R" "A"
[1463] "B" "B" "A" "B" "R" "B" "B" "B" "B" "R" "R" "A" "B" "P" "A" "A" "R"
[1480] "A" "P" "B" "P" "B" "P" "A" "A" "R" "A" "A" "R" "A" "P" "R" "B" "B"
[1497] "A" "A" "R" "R" "R" "R" "P" "P" "R" "P" "R" "R" "A" "A" "A" "R" "B"
[1514] "R" "A" "P" "R" "B" "A" "P" "P" "R" "P" "P" "R" "P" "P" "B" "P" "P"
[1531] "A" "R" "B" "B" "R" "A" "B" "R" "A" "P" "P" "P" "R" "P" "A" "R" "B"
[1548] "P" "P" "A" "R" "A" "A" "B" "A" "B" "R" "A"nrow(lettersAllTest)[1] 1558table(lettersAllTest$letter, lettersPredict) lettersPredict
A B P R
A 348 4 0 43
B 8 318 12 45
P 2 21 363 15
R 10 24 5 340(348 + 318 + 363 + 340) / nrow(lettersAllTest)[1] 0.8786906Problem 2.3 - Predicting the letters A, B, P, R
Now build a random forest model on the training data, using the same independent variables as in the previous problem – again, don’t forget to remove the isB variable.
Just use the default parameter values for ntree and node size (you don’t need to include these arguments at all). Set the seed to 1000 right before building our model. (Remember that you might get a slightly different result even if you set the random seed.)
set.seed(1000)
lettersForest <- randomForest(letter ~ . - isB, data = lettersAllTrain)What is the test-set accuracy of your random forest model?
lettersForestPredict <-
as.vector(predict(lettersForest, newdata = lettersAllTest, type = "class"))
lettersForestPredict [1] "B" "R" "A" "P" "A" "R" "A" "A" "A" "B" "A" "A" "P" "P" "B" "R" "B"
[18] "P" "B" "B" "A" "A" "P" "P" "R" "A" "A" "A" "B" "B" "A" "B" "P" "B"
[35] "B" "A" "P" "B" "B" "B" "A" "B" "R" "B" "R" "B" "R" "R" "P" "R" "P"
[52] "B" "R" "R" "B" "B" "R" "A" "B" "P" "R" "P" "B" "R" "B" "A" "P" "P"
[69] "R" "R" "A" "P" "P" "R" "B" "R" "A" "P" "R" "R" "P" "R" "B" "P" "A"
[86] "P" "A" "A" "A" "P" "A" "A" "A" "B" "B" "B" "R" "R" "P" "P" "B" "A"
[103] "R" "P" "R" "B" "B" "A" "P" "P" "R" "P" "A" "A" "P" "R" "P" "P" "P"
[120] "B" "R" "R" "A" "B" "R" "R" "P" "A" "B" "P" "P" "P" "B" "R" "A" "P"
[137] "P" "B" "B" "R" "A" "A" "B" "A" "P" "R" "R" "A" "R" "A" "B" "B" "B"
[154] "B" "B" "P" "A" "P" "B" "R" "A" "A" "P" "A" "A" "P" "B" "B" "B" "P"
[171] "P" "B" "R" "B" "R" "P" "P" "P" "B" "A" "P" "A" "A" "B" "R" "P" "P"
[188] "P" "R" "A" "B" "B" "A" "R" "B" "R" "P" "R" "A" "R" "B" "A" "R" "R"
[205] "B" "R" "B" "A" "B" "P" "A" "A" "A" "R" "B" "B" "A" "B" "P" "R" "R"
[222] "B" "P" "B" "A" "A" "B" "B" "B" "P" "R" "P" "A" "A" "A" "R" "A" "A"
[239] "B" "P" "R" "R" "A" "R" "A" "P" "B" "R" "P" "B" "A" "A" "B" "A" "A"
[256] "A" "B" "P" "A" "P" "A" "R" "P" "R" "A" "R" "P" "B" "P" "R" "P" "A"
[273] "R" "R" "R" "B" "R" "B" "P" "R" "P" "P" "A" "R" "P" "R" "B" "A" "P"
[290] "B" "A" "A" "A" "A" "B" "R" "R" "A" "P" "R" "B" "P" "B" "B" "R" "R"
[307] "R" "A" "B" "R" "R" "P" "R" "B" "A" "A" "P" "R" "A" "R" "P" "A" "R"
[324] "P" "R" "P" "P" "A" "P" "A" "A" "A" "A" "B" "P" "R" "P" "R" "P" "B"
[341] "B" "B" "A" "A" "B" "B" "B" "P" "B" "P" "B" "B" "A" "P" "R" "P" "R"
[358] "R" "P" "B" "P" "B" "P" "R" "B" "P" "B" "B" "P" "A" "P" "R" "A" "P"
[375] "B" "B" "R" "A" "B" "A" "R" "R" "A" "B" "P" "R" "A" "P" "P" "A" "A"
[392] "A" "P" "R" "R" "P" "B" "A" "A" "R" "P" "A" "R" "B" "B" "P" "P" "P"
[409] "B" "A" "R" "P" "A" "P" "A" "A" "A" "P" "P" "R" "R" "R" "B" "R" "P"
[426] "B" "B" "R" "B" "P" "P" "A" "A" "B" "A" "P" "P" "B" "P" "A" "R" "B"
[443] "B" "B" "R" "P" "A" "A" "B" "B" "P" "R" "A" "P" "A" "R" "P" "P" "P"
[460] "R" "B" "B" "P" "P" "P" "B" "R" "B" "P" "R" "B" "A" "R" "B" "P" "R"
[477] "R" "R" "P" "B" "B" "B" "A" "P" "B" "P" "P" "B" "A" "A" "A" "B" "P"
[494] "P" "R" "B" "A" "A" "B" "A" "R" "R" "R" "B" "R" "B" "R" "R" "R" "A"
[511] "A" "P" "R" "P" "A" "R" "B" "A" "P" "P" "R" "P" "R" "B" "A" "A" "P"
[528] "A" "B" "P" "A" "P" "A" "R" "A" "A" "R" "R" "P" "R" "A" "R" "R" "R"
[545] "B" "B" "R" "R" "B" "R" "A" "B" "A" "P" "B" "A" "A" "R" "P" "A" "A"
[562] "R" "R" "P" "A" "P" "R" "A" "R" "R" "A" "B" "R" "B" "B" "A" "R" "P"
[579] "B" "R" "B" "R" "P" "B" "A" "A" "B" "R" "A" "A" "B" "P" "P" "B" "P"
[596] "B" "P" "R" "A" "P" "P" "B" "B" "P" "B" "B" "P" "P" "P" "B" "A" "A"
[613] "B" "P" "B" "B" "A" "B" "A" "B" "A" "P" "R" "R" "A" "P" "B" "A" "P"
[630] "A" "P" "R" "R" "P" "B" "R" "A" "A" "A" "B" "B" "P" "B" "A" "P" "P"
[647] "P" "B" "A" "P" "P" "R" "P" "P" "A" "R" "B" "R" "B" "P" "B" "P" "P"
[664] "A" "P" "A" "R" "P" "P" "R" "A" "R" "P" "A" "B" "B" "R" "P" "P" "R"
[681] "B" "P" "R" "P" "R" "P" "A" "A" "A" "R" "A" "P" "R" "A" "B" "P" "P"
[698] "A" "B" "R" "R" "R" "A" "P" "A" "B" "R" "P" "B" "R" "P" "B" "P" "P"
[715] "A" "B" "P" "R" "B" "R" "B" "R" "B" "B" "B" "P" "A" "A" "R" "A" "A"
[732] "P" "R" "P" "P" "P" "P" "P" "R" "P" "R" "A" "P" "P" "B" "B" "B" "A"
[749] "B" "B" "R" "R" "B" "B" "R" "A" "B" "P" "P" "R" "A" "R" "P" "B" "B"
[766] "P" "P" "B" "B" "R" "B" "P" "B" "A" "A" "B" "A" "P" "A" "R" "P" "B"
[783] "R" "R" "A" "R" "P" "P" "P" "R" "A" "B" "B" "B" "B" "B" "A" "A" "P"
[800] "A" "B" "P" "R" "B" "B" "B" "P" "B" "P" "R" "A" "A" "B" "R" "B" "A"
[817] "P" "P" "P" "P" "B" "R" "P" "A" "R" "A" "A" "R" "R" "P" "P" "R" "R"
[834] "A" "P" "P" "B" "R" "P" "B" "B" "A" "R" "A" "B" "B" "B" "A" "P" "B"
[851] "R" "P" "A" "P" "A" "A" "B" "A" "P" "R" "B" "B" "B" "B" "R" "P" "A"
[868] "P" "R" "R" "P" "R" "R" "B" "P" "A" "B" "B" "P" "R" "R" "B" "B" "B"
[885] "A" "R" "A" "A" "A" "R" "R" "B" "R" "B" "P" "B" "A" "B" "B" "B" "R"
[902] "P" "P" "P" "B" "B" "R" "A" "A" "B" "P" "B" "A" "P" "P" "R" "R" "A"
[919] "R" "P" "R" "P" "R" "B" "R" "B" "R" "P" "P" "B" "A" "R" "B" "A" "R"
[936] "B" "P" "A" "A" "B" "P" "A" "R" "A" "P" "R" "B" "A" "A" "P" "A" "R"
[953] "P" "R" "B" "B" "A" "A" "P" "B" "B" "B" "A" "P" "P" "B" "A" "R" "A"
[970] "P" "P" "R" "P" "B" "R" "A" "B" "R" "P" "P" "B" "A" "A" "P" "B" "B"
[987] "A" "A" "A" "A" "A" "R" "A" "P" "P" "A" "A" "A" "B" "B" "B" "B" "R"
[1004] "P" "A" "P" "B" "B" "B" "A" "B" "A" "P" "R" "A" "P" "R" "P" "A" "R"
[1021] "P" "A" "B" "R" "P" "R" "A" "P" "P" "P" "P" "B" "R" "A" "R" "P" "A"
[1038] "R" "A" "B" "A" "P" "A" "R" "A" "A" "R" "B" "P" "A" "B" "B" "B" "B"
[1055] "B" "A" "A" "B" "B" "R" "R" "R" "B" "P" "B" "P" "A" "R" "A" "A" "P"
[1072] "R" "B" "A" "P" "R" "R" "A" "A" "P" "R" "R" "R" "R" "R" "A" "R" "B"
[1089] "P" "B" "B" "B" "P" "B" "B" "R" "A" "B" "A" "P" "R" "A" "B" "B" "A"
[1106] "B" "B" "P" "B" "B" "R" "P" "P" "A" "P" "A" "B" "R" "A" "A" "R" "R"
[1123] "B" "A" "P" "B" "B" "A" "A" "B" "B" "A" "R" "P" "B" "P" "B" "B" "A"
[1140] "A" "P" "R" "R" "A" "R" "B" "P" "R" "A" "R" "R" "P" "B" "P" "R" "B"
[1157] "P" "A" "A" "P" "R" "R" "A" "R" "B" "R" "P" "A" "B" "P" "R" "B" "B"
[1174] "A" "A" "A" "R" "B" "R" "B" "A" "R" "A" "B" "R" "R" "R" "A" "A" "B"
[1191] "B" "B" "P" "P" "B" "R" "P" "A" "A" "A" "B" "P" "A" "P" "R" "A" "P"
[1208] "P" "P" "R" "R" "R" "P" "P" "R" "A" "P" "R" "B" "A" "B" "R" "B" "A"
[1225] "A" "P" "R" "R" "B" "A" "A" "P" "R" "P" "A" "A" "A" "B" "A" "P" "A"
[1242] "B" "B" "P" "B" "B" "R" "A" "P" "R" "B" "R" "P" "B" "A" "A" "R" "P"
[1259] "B" "A" "R" "R" "A" "R" "P" "P" "B" "A" "P" "R" "B" "A" "A" "P" "A"
[1276] "B" "R" "P" "P" "A" "P" "P" "B" "A" "A" "R" "P" "R" "A" "P" "R" "A"
[1293] "P" "R" "A" "P" "P" "A" "R" "B" "B" "R" "R" "B" "B" "R" "R" "A" "B"
[1310] "A" "R" "R" "P" "A" "R" "R" "A" "R" "B" "R" "B" "R" "R" "A" "A" "B"
[1327] "B" "P" "R" "R" "P" "R" "B" "R" "B" "A" "R" "A" "A" "A" "R" "B" "P"
[1344] "P" "P" "B" "B" "A" "P" "R" "B" "R" "P" "P" "P" "A" "R" "B" "A" "A"
[1361] "B" "B" "A" "P" "R" "B" "P" "P" "R" "A" "B" "R" "P" "B" "B" "A" "A"
[1378] "R" "B" "P" "P" "R" "P" "P" "P" "P" "A" "B" "R" "B" "A" "B" "R" "P"
[1395] "B" "P" "A" "A" "P" "P" "A" "B" "R" "P" "B" "R" "A" "P" "R" "B" "R"
[1412] "A" "B" "B" "R" "P" "P" "P" "A" "B" "A" "A" "A" "P" "P" "A" "B" "B"
[1429] "P" "B" "B" "R" "B" "A" "P" "B" "R" "R" "A" "P" "P" "P" "P" "P" "R"
[1446] "A" "B" "A" "B" "A" "B" "B" "P" "B" "A" "B" "P" "R" "P" "P" "A" "A"
[1463] "B" "B" "A" "B" "R" "B" "B" "P" "B" "R" "R" "A" "B" "P" "A" "A" "R"
[1480] "A" "P" "B" "P" "B" "P" "A" "R" "R" "A" "A" "R" "A" "P" "B" "B" "B"
[1497] "A" "A" "R" "R" "R" "R" "P" "P" "B" "P" "R" "B" "A" "A" "A" "R" "R"
[1514] "R" "A" "P" "R" "B" "A" "P" "P" "A" "P" "P" "R" "P" "P" "B" "P" "P"
[1531] "A" "R" "B" "B" "P" "R" "B" "R" "A" "P" "P" "P" "B" "P" "A" "R" "B"
[1548] "P" "P" "A" "R" "A" "A" "B" "A" "P" "R" "A"table(lettersAllTest$letter, lettersForestPredict) lettersForestPredict
A B P R
A 391 0 3 1
B 0 380 1 2
P 0 6 394 1
R 3 14 0 362(390 + 380 + 393 + 364) / nrow(lettersAllTest)[1] 0.9801027Conclusion
You should find this value rather striking, for several reasons. The first is that it is significantly higher than the value for CART, highlighting the gain in accuracy that is possible from using random forest models.
The second is that while the accuracy of CART decreased significantly as we transitioned from the problem of predicting B or not B (a relatively simple problem) to the problem of predicting the four letters (certainly a harder problem), the accuracy of the random forest model decreased by a tiny amount.
